
RESEARCH
eco-evolutionary community dynamics


In our work, the main focus has been to understand what drives the dynamics in ecological communities. We are especially interested in how contemporary rapid evolution alters ecological dynamics and how ecological dynamics, in turn, feed back to evolutionary processes.
Currently, the field moves beyond observations in simple environments and simple few-species setting asking questions about the effect of temporally and spatially varying selection pressures and community complexity. Increasing complexity at any level, however, results in a number of challenges that often can only be tackled using a combination of experimental evolution, mathematical modelling and molecular techniques. Currently our focus is on testing eco-evolutionary feedbacks in species-rich microbial communities to see if the finding from simple few-species systems still apply in the presence of more complex species interactions. Another theme is to study how environmental factors, such as sublethal antibiotic concentrations, alter eco-evolutionary dynamics.
Evolution of antibiotic resistance in the presence of ecological interactions

Controlled studies on factors influencing antibiotic resistance mutational trajectories and horizontal transfer of antibiotic resistance genes have primarily focused on single bacterial species. However, bacteria do not live alone but are embedded in complex multispecies, multi-trophic communities. This can affect fundamental features of bacterial ecology and evolution, with both direct and indirect effects on antibiotic resistance trajectories. We combine our expertise on community ecology and experimental evolution to investigate antibiotic resistance in microbial communities in detail. Alongside tracking fitness-related phenotypic traits of interest over time in our experiments, we track molecular factors using high-throughput sequencing methods, including both amplicon-based and omics approaches.
Host-parasite interaction between Baltic Sea cyanobacteria and their phages
One line of our research concentrates on more applied questions using harmful Baltic Sea cyanobacteria as a model organism. In this project, we investigate the ecological and evolutionary role of cyanobacterial viruses in cyanobacterial population dynamics and bloom formation. Here we apply recent eco-evolutionary theory to the relatively well-studied field of harmful cyanobacteria, with a focus on the Baltic Sea where cyanobacteria have wide socioeconomic impacts.

In filamentous cyanobacteria gaseous nitrogen (N2) is fixed in heterocysts and distributed to the vegetative cells along the filament when extracellular inorganic nitrogen is scarce. Part of the fixed nitrogen is utilized to build cellular structures while part of it will leak out from the cells as dissolved inorganic nitrogen (DIN) or dissolved organic nitrogen (DON). DON can then be transformed into DIN by bacteria (remineralization) becoming thus available for reuse. In addition, DIN, DON and particulate organic nitrogen (PON) are released from decaying or lysed cells. The fixed nitrogen may be transferred not only to other members of the bacterioplankton and phytoplankton community but also to primary consumers such as zooplankton while they graze on fresh, decaying or phage-infected cyanobacteria.
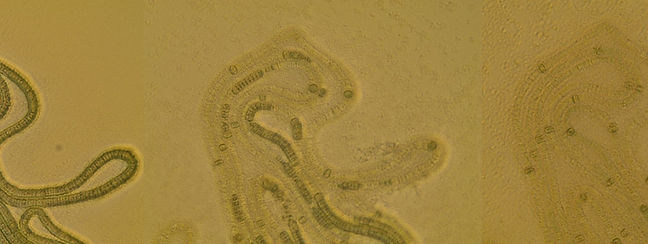
Filamentous cyanobacteria Nodularia spumigena before and after phage infection.